In order to fully understand the biology of Alzheimer’s disease, create new predictive models and identify novel causal and druggable targets large scale unbiased omic analyses are needed.
For the last three years my lab has embarked on generated the largest and most detailed proteomic atlas in brain, CSF and plasma from Alzheimer’s, Parkinson’s disease and other neurodegenerative diseases.
A good example of how this data will lead to a better understanding of Alzheimer’s disease is a recent published paper (Proteo-genomics of soluble TREM2 in cerebrospinal fluid provides novel insights and identifies novel modulators for Alzheimer’s disease | Molecular Neurodegeneration | Full Text (biomedcentral.com)) in where we integrated genetics and proteomic data to identify additional genes and proteins that are part of the TREM2 pathway. By performing multi-ethnic mapping, we were able to identify the two functional variants on the MS4A locus. In top of that, we identified and functionally characterize two novel loci: TGFB2 and Nectin2. These findings were possible by analyzing large human proteomic datasets, as it is unclear whether the current cell and animal models would have captured these new genes and protein-protein interactions.
Building on that, we generated the largest CSF proteomic dataset in terms of samples (more than 3,000 samples) and proteins (more than 7,000 proteins) from sporadic (Multi-cohort cerebrospinal fluid proteomics identifies robust molecular signatures for asymptomatic and symptomatic Alzheimer’s disease. | Research Square) and autosomal dominant AD (Systematic proteomics in Autosomal dominant Alzheimer’s disease reveals decades-early changes of CSF proteins in neuronal death, and immune pathways | medRxiv).
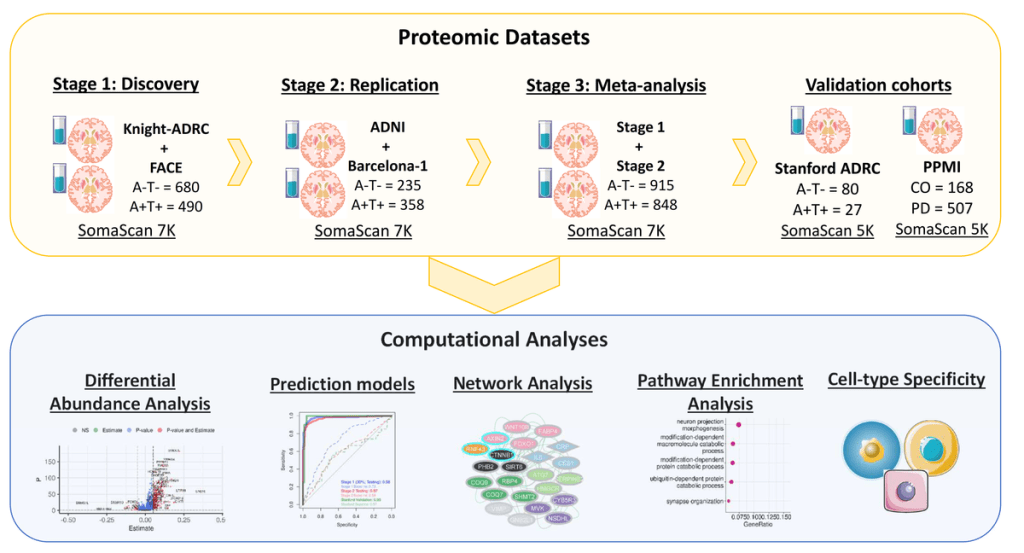
In these two preprints we identified and replicated more than 2,000 proteins associated with clinical and biomarker status in sporadic AD and more than 120 for autosomal dominant AD. We later leveraged those proteins to create novel predictive models as well as to identify pathways associated with disease.
In these two studies we were able to generate, replicate and validated novel predictive models that include a small number of proteins that predict AD with extremely high power. This is important as those novel predictive models are Aβ and tau-independent. As current therapies are targeting Aβ and tau, those proteins cannot be used as biomarkers but for target engagement. Our study present novel biomarkers that could be used to track disease status in individuals taking new therapies.
In addition, in our study looking at sporadic AD, and because the large number of individuals and proteins, we were able to demonstrate that the proteins associated with AD, follow four unique trajectories capturing different biological processes, including 1) death of glutamatergic and dopaminergic neurons, 2) activation of immune response and endolysosomal dysfunction, 3) brain plasticity and mechanism to compensate for AD-related pathology and 4) proteins involved on cell-to-cell crosstalk including microglia and astrocytes
Multi-cohort cerebrospinal fluid proteomics identifies robust molecular signatures for asymptomatic and symptomatic Alzheimer’s disease
Changes in the Amyloid-β (Aβ) and hyperphosphorylated Tau proteins in brain and cerebrospinal fluid (CSF) precedes AD symptoms, making CSF proteome a potential avenue to understand the pathophysiology and facilitate reliable diagnostics and therapies. While CSF and plasma Aβ and Tau levels are considered the gold standard biomarkers for AD, additional models are needed as most of the current disease-modifying therapies in clinical trials target Aβ and/or tau with a limited outcome. In order to identify novel Aβ- and Tau-independent proteomics biomarkers for AD, we measured and analyzed the protein expression levels of 7,029 analytes in CSF of 2,286 participants from multi-center cohorts. We employed a three-stage analytical approach (discovery, replication, and meta-analysis) to identify robust AD CSF proteomic alterations. Machine learning was implemented to create and validate highly accurate and replicable 11-protein model that predict classical AD biomarker positivity and clinical status. The predicted model is AD-specific as it does not perform well for other neurodegenerative disorders (e.g., Parkinson’s disease, Frontotemporal dementia, and dementia with Lew bodies) and it can also identify people that will convert to AD as well as those AD cases with faster progression.
Systematic proteomics in Autosomal dominant Alzheimer’s disease reveals decades-early changes of CSF proteins in neuronal death, and immune pathways
This study advances proteomic research in Autosomal Dominant Alzheimer’s Disease (ADAD) by employing an unprecedentedly comprehensive proteomic panel (more than 7,000 protein analytes) to analyze cerebrospinal fluid (CSF) and plasma in 291 mutation carriers (MCs) and 185 non-carriers (NCs), alongside data from four sporadic late-onset Alzheimer’s disease (sAD) cohorts totaling over 1,700 participants. Unlike prior work, this research innovates by assessing pseudo-trajectories based on the estimated age at onset, thus simulating longitudinal data from cross-sectional observations to highlight proteins with divergent trajectories between MCs and NCs. This method determined the critical juncture of divergence in disease progression and further replicated our findings using publicly accessible ADAD datasets. Additionally, co-expression network analysis identified protein modules linked to different disease stages, revealing insights into stress response, neuronal death, and cell-to-cell communication changes. Leveraging machine learning, we distilled a nine-protein signature with superior predictive capability for ADAD carrier status than existing biomarkers, independent of Aβ and tau, offering a fresh avenue for prediction models. These novel methodologies and insights provide a deeper understanding of ADAD progression and hold significant implications for the scientific community.
This study was recently covered at Alzforum:. Proteomics Uncovers Potential Markers of Early Autosomal Dominant AD | ALZFORUM