Transposable elements have been implicated in neurodegeration, including Alzheimer disease (AD). Recent genetic studies have highlighted the role of not only neurons but also microglia in the pathogenesis of AD. Therefore, to fully understand the biology of the disease there is a need to perform depth molecular characterization in a cell-specific manner.
We and others have shown that AD is associated with changes in brain cell proportion and transcriptomic changes, some of them are also cell specific. Additionally, the latest genetic studies implicate cell-specific pathogenic events that lead to disease. Pathogenic variants in APP, PSEN1 and PSEN2 affects APP processing leading to Aβ aggregates and neuronal death. Genetic variants in TREM2 and MS4A modify AD risk by affecting microglia activity. To fully understand and characterize the role of transposable elements (TE) in AD pathogenesis there is a need to novel and multidisciplinary approaches. In this project we will combine novel genomic approaches in human brain tissues, direct converted neurons and iPSC-derived microglia (iMGL) to identify cell-specific TE and downstream (chromatin accessibility, transcription) changes implicated in AD. We will leverage a large and unique resources of human brain samples and fibroblast from individuals with mutations in APP, PSEN1, PSEN2, as well as risk variants in TREM2, MS4A or APOE. We will also use the direct converted neurons and iMGL, together with new genomic editing approaches to target and characterize the mechanism by which TE contribute to disease.
The overall goal of this proposal is to identify and characterize neuronal and microglia-specific TE and downstream (chromatin accessibility, transcript levels and new transcript) changes implicated on disease. Functional characterization of TE implicated in disease will also be instrumental to identify potential therapeutic targets.
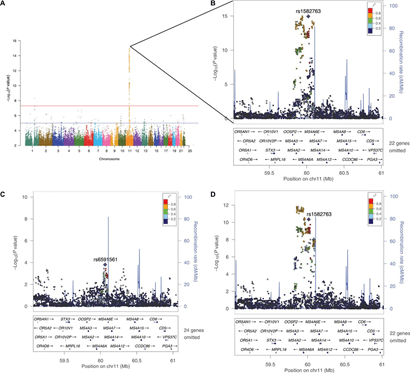
The MS4A gene cluster is a key modulator of soluble TREM2 and Alzheimer’s disease risk. (A) Manhattan plot shows the negative log10-transformed P values on the y axis for CSF sTREM2 concentrations based on measurements of ADNI CSF samples by Ludwig-Maximilians-Universität. (B) Regional association plot of the MS4A gene region in the single-variant analysis. (C) Regional association plot of the MS4A region after conditioning on the top SNP (rs1582763). (D) Regional association plot of the MS4A region after conditioning on SNP rs6591561 (MS4A4A p.M159V). The SNPs for each regional plot are represented by a purple diamond. Each dot represents individual SNPs, and dot colors in the regional plots represent linkage disequilibrium with the named SNP. Blue vertical lines in the regional plots show recombination rate as marked on the right-hand y axis.
Publications
- Novotny BC, Fernandez MV, Wang C, Budde JP, Bergmann K, Eteleeb AM, Bradley J, Webster C, Ebl C, Norton J, Gentsch J, Dube U, Wang F, Morris JC, Bateman RJ, Perrin RJ, McDade E, Xiong C, Chhatwal J; Dominantly Inherited Alzheimer Network (DIAN) Study Group; Alzheimer’s Disease Neuroimaging Initiative; and the Alzheimer’s Disease Metabolomics Consortium (ADMC), Goate A, Farlow M, Schofield P, Chui H, Karch CM, Cruchaga C, Benitez BA, Harari O. Metabolomic and lipidomic signatures in autosomal dominant and late-onset Alzheimer’s disease brains. Alzheimers Dement. 2022 Oct 17. doi: 10.1002/alz.12800. Epub ahead of print. PMID: 36251323.
- Novotny BC, Fernandez MV, Wang C, Budde JP, Bergmann K, Eteleeb AM, Bradley J, Webster C, Ebl C, Norton J, Gentsch J, Dube U, Wang F, Morris JC, Bateman RJ, Perrin RJ, McDade E, Xiong C, Chhatwal J; Dominantly Inherited Alzheimer Network (DIAN) Study Group; Alzheimer’s Disease Neuroimaging Initiative; and the Alzheimer’s Disease Metabolomics Consortium (ADMC), Goate A, Farlow M, Schofield P, Chui H, Karch CM, Cruchaga C, Benitez BA, Harari O. Metabolomic and lipidomic signatures in autosomal dominant and late-onset Alzheimer’s disease brains. Alzheimers Dement. 2022 Oct 17. doi: 10.1002/alz.12800. Epub ahead of print. PMID: 36251323.
- Olive C, Ibanez L, Farias FHG, Wang F, Budde JP, Norton JB, Gentsch J, Morris JC, Li Z, Dube U, Del-Aguila J, Bergmann K, Bradley J, Benitez BA, Harari O, Fagan A, Ances B, Cruchaga C, Fernandez MV. Examination of the Effect of Rare Variants in TREM2, ABI3, and PLCG2 in LOAD Through Multiple Phenotypes. J Alzheimers Dis. 2020;77(4):1469-1482. doi: 10.3233/JAD-200019. PMID: 32894242; PMCID: PMC7927150.
- Cignarella F, Filipello F, Bollman B, Cantoni C, Locca A, Mikesell R, Manis M, Ibrahim A, Deng L, Benitez BA, Cruchaga C, Licastro D, Mihindukulasuriya K, Harari O, Buckland M, Holtzman DM, Rosenthal A, Schwabe T, Tassi I, Piccio L. TREM2 activation on microglia promotes myelin debris clearance and remyelination in a model of multiple sclerosis. Acta Neuropathol. 2020 Oct;140(4):513-534. doi: 10.1007/s00401-020-02193-z. Epub 2020 Aug 9. PMID: 32772264; PMCID: PMC7498497.
- Deming Y, Filipello F, Cignarella F, Cantoni C, Hsu S, Mikesell R, Li Z, Del-Aguila JL, Dube U, Farias FG, Bradley J, Budde J, Ibanez L, Fernandez MV, Blennow K, Zetterberg H, Heslegrave A, Johansson PM, Svensson J, Nellgård B, Lleo A, Alcolea D, Clarimon J, Rami L, Molinuevo JL, Suárez-Calvet M, Morenas-Rodríguez E, Kleinberger G, Ewers M, Harari O, Haass C, Brett TJ, Benitez BA, Karch CM, Piccio L, Cruchaga C. The MS4A gene cluster is a key modulator of soluble TREM2 and Alzheimer’s disease risk. Sci Transl Med. 2019 Aug 14;11(505):eaau2291. doi: 10.1126/scitranslmed.aau2291. PMID: 31413141; PMCID: PMC6697053.
- Del-Aguila JL, Benitez BA, Li Z, Dube U, Mihindukulasuriya KA, Budde JP, Farias FHG, Fernández MV, Ibanez L, Jiang S, Perrin RJ, Cairns NJ, Morris JC, Harari O, Cruchaga C. TREM2 brain transcript-specific studies in AD and TREM2 mutation carriers. Mol Neurodegener. 2019 May 8;14(1):18. doi: 10.1186/s13024-019-0319-3. PMID: 31068200; PMCID: PMC6505298.
- Deming Y, Li Z, Benitez BA, Cruchaga C. Triggering receptor expressed on myeloid cells 2 (TREM2): a potential therapeutic target for Alzheimer disease? Expert Opin Ther Targets. 2018 Jul;22(7):587-598. doi: 10.1080/14728222.2018.1486823. Epub 2018 Jun 20. PMID: 29889572; PMCID: PMC6295143.