Circular RNAs (circRNAs) are a class of RNAs that result from backsplicing events, in which the 3′-ends of transcripts are covalently spliced with the 5′-ends, thereby forming continuous loops. As RNA-seq has become widespread, thousands of circRNAs have been identified across eukaryotes. These studies have found circRNAs to be highly expressed in the nervous system and enriched in synapses. In the brain, circRNA expression can occur independently of linear transcript expression, and may be a gene’s most highly expressed isoform. Brain circRNAs are also regulated during development and in response to neuronal excitation. CircRNAs accumulate in aging mouse and fly brains, possibly due to their lack of free hydroxyl ends conferring resistance to exonucleases. Much is still unknown about circRNA biology; for example, it was only recently demonstrated that circRNAs can be translated in vivo. Thus far, the most well-established role of circRNAs is in miRNA regulation via sequestration, leading to loss of function.
Alzheimer’s disease (AD) is a progressive, neurodegenerative disorder and the most common cause of dementia, affecting millions worldwide. It is neuropathologically characterized by the accumulation of amyloid beta plaques and tau inclusions, as well as widespread neuronal atrophy, which results in dramatic cognitive impairment. Unfortunately, no effective preventive, palliative or curative therapies currently exist for AD. Previous studies investigating linear transcriptomic (mainly messenger RNA) differences in the context of AD have yielded insight into the pathogenic mechanisms underlying this disease, as well as potential therapeutic targets. Similar analyses for circRNAs remain outstanding, although a single circRNA that regulates specific miRNAs and synaptic function—CDR1-AS—has been reported to be downregulated in AD brains.
This project will use brain tissue to identify additional brain circRNAs implicated in AD from a cohort that is four-fold larger than the cohort in our previous study. We also plan to investigate differentially expressed blood circRNAs in AD cases compared with controls to determine their biomarker utility for creating new prediction models. We will establish a framework for in vitro and in vivo functional characterization of the role of circRNAs in AD. As proof of principle, we will start by defining the role of circHOMER1 in AD-related gene expression and related cellular phenotypes. We have found that circHOMER1 is highly expressed in induced pluripotent stem cell (iPSC)-derived neurons. We will use CRISPR/Cas9 to knock down circHOMER1 as well as use circHOMER1 overexpression (OE) in iPSC-derived neurons from isogenic controls and AD patient-derived neuronal cultures from pathogenic mutation carriers of APP, PSEN1, PSEN2, and MAPT genes. To determine whether circHomer1 accelerate/increase AD-related pathology in vivo, we will generate circHomer1-knockout transgenic mice and cross them with 5XFAD and MAPT-P301S mice. We will also use AAV2/9-mediated circHomer1 OE in the cortex and hippocampus of 5XFAD and MAPT-P301S mice. We recently found that seven-month-old 5XFAD mice display significant reductions of circHomer1 expression, similar to the postmortem brains of AD patients. We will restore circHomer1 levels in the hippocampus and cortex of 5XFAD and P301S mice using AAV2/9 vectors. This proposal will be the first to systematically analyze the role of brain and blood circRNAs in AD and to perform in vitro and in vivo functional studies to characterize the role of circRNAs in AD
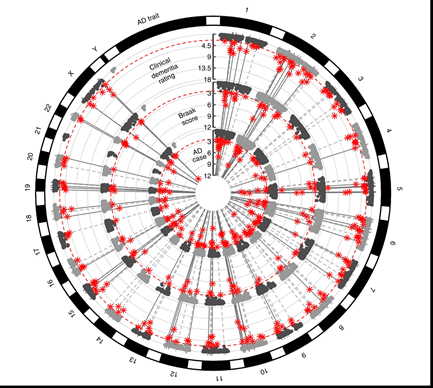
Each circular Manhattan plot presents the results from a meta-analysis of circRNA AD-association results from discovery (parietal cortex) and replication (inferior frontal gyrus (Brodmann Area 44)) datasets. In order from outermost to innermost circular plot, the AD traits include: clinical dementia rating at expiration/death (CDR), Braak neuropathological severity score, and AD case-control status (AD case). Study-wide significance threshold is based on a false discovery rate of 0.05 and depicted by the red, dashed line. circRNAs that passed this threshold are displayed with star symbols. Lines extending through all three plots identify circRNAs that are significantly associated with multiple AD traits.
Publications
- Dube U, Del-Aguila JL, Li Z, Budde JP, Jiang S, Hsu S, Ibanez L, Fernandez MV, Farias F, Norton J, Gentsch J, Wang F; Dominantly Inherited Alzheimer Network (DIAN), Salloway S, Masters CL, Lee JH, Graff-Radford NR, Chhatwal JP, Bateman RJ, Morris JC, Karch CM, Harari O, Cruchaga C. An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations. Nat Neurosci. 2019 Nov;22(11):1903-1912. doi: 10.1038/s41593-019-0501-5. Epub 2019 Oct 7. PMID: 31591557; PMCID: PMC6858549.
- Chen HH, Eteleeb A, Wang C, Fernandez MV, Budde JP, Bergmann K, Norton J, Wang F, Ebl C, Morris JC, Perrin RJ, Bateman RJ, McDade E, Xiong C, Goate A, Farlow M, Chhatwal J, Schofield PR, Chui H, Harari O, Cruchaga C, Ibanez L; Dominantly Inherited Alzheimer Network. Circular RNA detection identifies circPSEN1 alterations in brain specific to autosomal dominant Alzheimer’s disease. Acta Neuropathol Commun. 2022 Mar 4;10(1):29. doi: 10.1186/s40478-022-01328-5. PMID: 35246267; PMCID: PMC8895634.